In-Silico discovery of 17alpha-hydroxywithanolide-D as potential neuroprotective allosteric modulator of NMDA receptor targeting Alzheimer's disease – PubMed Black Hawk Supplements
BLACK HAWK: High quality shilajit supplement for
Published article
Alzheimer’s disease (AD) is a progressive neurodegenerative disorder marked by cognitive decline, memory impairment, and behavioral alterations. The N-methyl-D-aspartate (NMDA) receptor has emerged as a promising target for AD pharmacotherapy due to its role in the disease’s pathogenesis. This study leverages advanced computational methods to screen 80 active constituents of Withania somnifera (Ashwagandha), a traditional herb known for its neuroprotective effects, against the NMDA receptor,…
Black Hawk Supplements, best supplements in the UK

In-Silico discovery of 17alpha-hydroxywithanolide-D as potential neuroprotective allosteric modulator of NMDA receptor targeting Alzheimer’s disease
Manoj Kumar Vashisth et al. Sci Rep. .
Abstract
Alzheimer’s disease (AD) is a progressive neurodegenerative disorder marked by cognitive decline, memory impairment, and behavioral alterations. The N-methyl-D-aspartate (NMDA) receptor has emerged as a promising target for AD pharmacotherapy due to its role in the disease’s pathogenesis. This study leverages advanced computational methods to screen 80 active constituents of Withania somnifera (Ashwagandha), a traditional herb known for its neuroprotective effects, against the NMDA receptor, using FDA-approved Ifenprodil as a reference. Our blind virtual screening results demonstrated that all tested compounds could bind to various domains of the NMDA receptor, with binding energies ranging from – 4.1 to -11.9 kcal/mol, compared to Ifenprodil’s -7.8 kcal/mol. Binding preference analysis revealed 7 compounds bound to the A-chain, 37 to the B-chain, 7 to the C-chain, and 29 to the D-chain of the receptor. Notable binding was observed predominantly at the Amino Terminal Domain (ATD) core site, some at the ATD-Ligand Binding Domain (LBD) interface, and a few at the Transmembrane Domain (TMD). Particularly, 17alpha-hydroxywithanolide D, with a binding energy of -11.9 kcal/mol, emerged as a prime candidate for further investigation. Molecular dynamics simulations of this compound revealed key interactions, including direct hydrogen bonding with residues ASP165, ARG431, THR433, LYS466, and TYR476 on the D-chain, as well as additional hydrophobic and water-bridging interactions. These simulations highlighted the compound’s influence on dynamic conformational states of the GluN1b-GluN2B receptor complex, modulating interactions between GluN1b Lys178 and GluN2B Asn184. Furthermore, the compound affected the distance between LBD heterodimers and the tension within the LBD-M30 linker, demonstrating its potential to modulate NMDA receptor activity. This comprehensive study not only underscores the therapeutic promise of Withania somnifera derivatives for AD but also provides a detailed molecular basis for their efficacy, offering valuable insights for targeted drug development and innovative therapeutic strategies against Alzheimer’s disease.
Keywords: Withania somnifera; 17alpha-hydroxywithanolide D; Alzheimer’s disease; Docking; Ifenprodil; Molecular dynamics; N-methyl-D-aspartate receptor; Virtual screening.
© 2024. The Author(s).
Conflict of interest statement
Figures

The three-dimensional structural representation of the NMDA receptor providing a detailed examination from various perspectives, each designated with specific colors for chain identification. In the front view, the A-chain stands out distinctly in a striking Red color, offering a clear visualization of its positioning. Transitioning to the top view, the B-chain takes center stage with its prominent Blue color, facilitating a comprehensive overhead perspective. The bottom view highlights the C-chain, uniquely designated in Green, aiding in a focused examination of its spatial arrangement. Completing the structural overview, the Yellow color specifically designates the D-chain, ensuring clarity in identifying its position within the overall structure.

The visual snapshot captures a moment in the dynamic interaction between the NMDA receptor chains and the virtually screened compound. Each receptor chain—A, B, C, and D—is depicted distinctly, showcasing the binding of the screened compound at various domains.
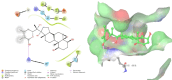
Molecular level interactions between NMDA–17alpha-hydroxywithanolide D complex. Left panel shows the interactions in 2D, whereas the right panel shows the interactions in three dimensional representation.
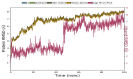
Root mean square deviation (RMSD) of protein and ligand backbone of NMDA-17alpha-hydroxywithanolide D complex. X-axis represents the timeline of the simulation in terms of nanoseconds, whereas the y-axis represents the calculated RMSD in angstrom units.
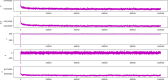
Calculated potential energies of the simulated systems of NMDA-17alpha-hydroxywithanolide D complex. X-axis represents the timeline of the simulation in terms of nanoseconds, whereas the y-axis represents the calculated potential energy in kcal/mol units.
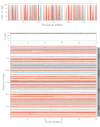
Analysis of Secondary Structural Elements (SSE) of NMDA-17alpha-hydroxywithanolide D compound complex. Protein secondary structure elements (SSE) like alpha-helices (orange colored) and beta-strands (blue colored) are monitored throughout the simulation. As delineated in the plot above, the X-axis corresponds to the amino acid residues index, spanning the entire protein structure. On the other hand, the y-axis represents the calculated SSE percentage concerning the simulated timescale. The distinctive coloring of alpha-helices and beta-strands aids in visually discerning their respective contributions to the overall secondary structure.
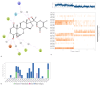
Comprehensive Analysis of Molecular Interactions in NMDA-17alpha-hydroxywithanolide D Complex: (a) Molecular Dynamics Simulations Interactions: During the extensive molecular dynamics simulations of the NMDA-17alpha-hydroxywithanolide D complex, intricate molecular-level interactions were observed. (b) Interaction Bar Chart Panel: The bar chart panel above provides a graphical representation of ligand-interacting amino acid residues on the x-axis and their respective interaction percentages concerning the simulated timescale on the y-axis. (c) Timeline Representation of Interactions and Contacts: The timeline representation encapsulates a comprehensive summary of various contacts, including H-bonds, hydrophobic interactions, ionic interactions, and water bridges.
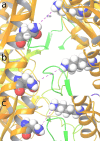
Visualization of NMDA subunit re-arrangement within GluN1b-GluN2B ATD Heterodimers during the simulated timescale from molecular dynamic simulations trajectory analysis. The configuration of subunits within the ATD heterodimers is delineated by the distance between GluN1b Lys178 and GluN2B Asn184 in the R2 lobes at (a) first frame, (b) 50ns time scale frame and (c) last frame of the simulation.
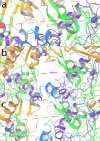
Visualization of NMDA relative orientation of GluN1b-GluN2B LBD heterodimers during the simulated timescale from molecular dynamic simulations trajectory analysis: The positioning of LBD heterodimers is defined by the distance between GluN1b Arg510 in loop1 (L10) and GluN2B Leu425 in loop2 (L2) in the D1 lobes, known as the L10-L2 distance at (a) first frame, (b) 50ns time scale frame and (c) last frame of the simulation.
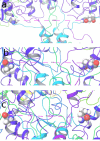
Visualization of relative tension of LBD-M30 linker of GluN2B subunits during the simulated timescale from molecular dynamic simulations trajectory analysis: The tension within the LBD-M30 linker of GluN2B subunits, a critical factor influencing the channel gate, is quantified by the distance between the two GluN2B Gln662 residues from the respective subunits at (a) first frame, (b) 50ns time scale frame and (c) last frame of the simulation.
References
-
- Trompetero, A. et al. Alzheimer’s disease and Parkinson’s disease: a review of current treatment adopting a nanotechnology approach. Curr. Pharm. Design. 24 (1), 22–45 (2018). – PubMed
-
- Alzheimer’s, A. 2017 Alzheimer’s disease facts and figures. Alzheimer’s Dement.13 (4), 325–373 (2017).
-
- Prince, M. et al. The global prevalence of dementia: a systematic review and metaanalysis. Alzheimer’s Dement.9 (1), 63–75 (2013). – PubMed
MeSH terms
Substances
Grants and funding
BLACK HAWK: Best ashwagandha supplement for mums
Read the original publication: